Multi-omics Data Analysis
Match Multi-omics Using Contrastive Pretraining With Feature Disentanglement
Project Background
In order to acquire knowledge in the field of multi-omics, I enrolled in a multi-omics artificial intelligence course in this semester. This project was the final assignment for the course. The course instructor believes that my idea for the final project can be further researched to produce meaningful results, so I am currently working diligently towards that goal.
Basic Idea
This project concentrate on learning modality shared information building model based on variational auto-encoders with feature disentanglement predicting one modality from another in the same cells as task1. Then I further to train a modality-shared information omics-content pretraining encoder to match multi-omics, following the paradigm of Contrastive Language-Image Pre-Training(CLIP).
Introduction of VAE
A Variational Autoencoder (VAE) \cite{vaetutorial} is a probabilistic spin on the classic autoencoder, a type of neural network used for unsupervised learning of efficient codings. Unlike a traditional autoencoder, which maps input data \(x\) to a hidden representation \(h\) and back to a reconstruction \(\hat{x}\) without considering how the hidden representation is structured, a VAE assumes that the data is generated by a directed graphical model \(p(x|z)\) and the encoder learns an approximation \(q_\phi(z|x)\) to the posterior distribution over the latent variables \(z\).
The architecture of a VAE consists of two main components: the encoder and the decoder. The encoder transforms the input data into two parameters in a latent space, which are \(\mu\) and \(\sigma\), representing the mean and standard deviation of a Gaussian distribution from which we sample the latent variable \(z\). This is expressed as:
\[z = \mu + \sigma \odot \epsilon\]where \(\epsilon\) is a random variable sampled from a standard normal distribution to provide stochasticity, and \(\odot\) represents element-wise multiplication.
The decoder then takes this latent variable \(z\) and reconstructs the input data \(x\). The objective function of a VAE is the Evidence Lower Bound (ELBO), which is given by:
\[\text{ELBO} = \mathbb{E}_{q_\phi(z|x)}[\log p_\theta(x|z)] - D_{KL}(q_\phi(z|x) \| p(z))\]Here, \(\theta\) and \(\phi\) are parameters of the decoder and encoder which are learned during training. \(D_{KL}\) denotes the Kullback-Leibler divergence between the encoder’s distribution \(q_\phi(z|x)\) and \(p(z)\), the prior distribution over the latent variables, which is typically assumed to be a standard normal distribution. The ELBO thus balances two objectives: the reconstruction fidelity, measured by the expected log-likelihood, and a regularization term, which keeps the learned distribution close to the prior.
Insight
It is apparent and intuitional that there is modality shared information or message flowing from DNA to RNA, ultimately to protein, according to the central dogma. Since that, it is plausible and feasible that it is possible to extract the core message from upstream modality, then translate the core message into the downstream modality. Hung-yi Lee etc, find that simply adding Instance normalization (IN) can remove the speaker information while preserving the content information. Similar idea has been verified to be effective for style transfer in computer vision. This is the original idea and I am endeavoring and collaborating with course instructor jingzhuo wang to further this idea.
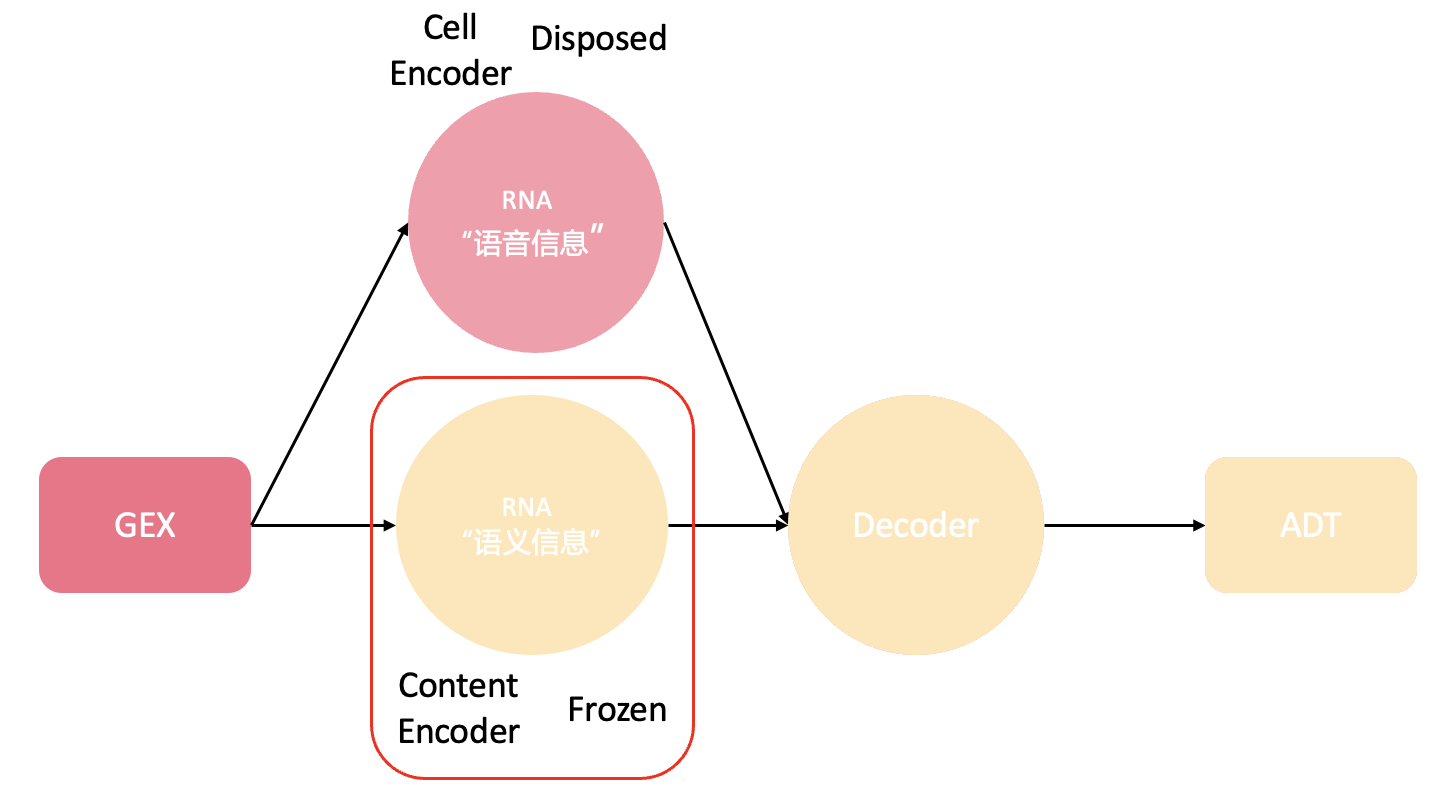
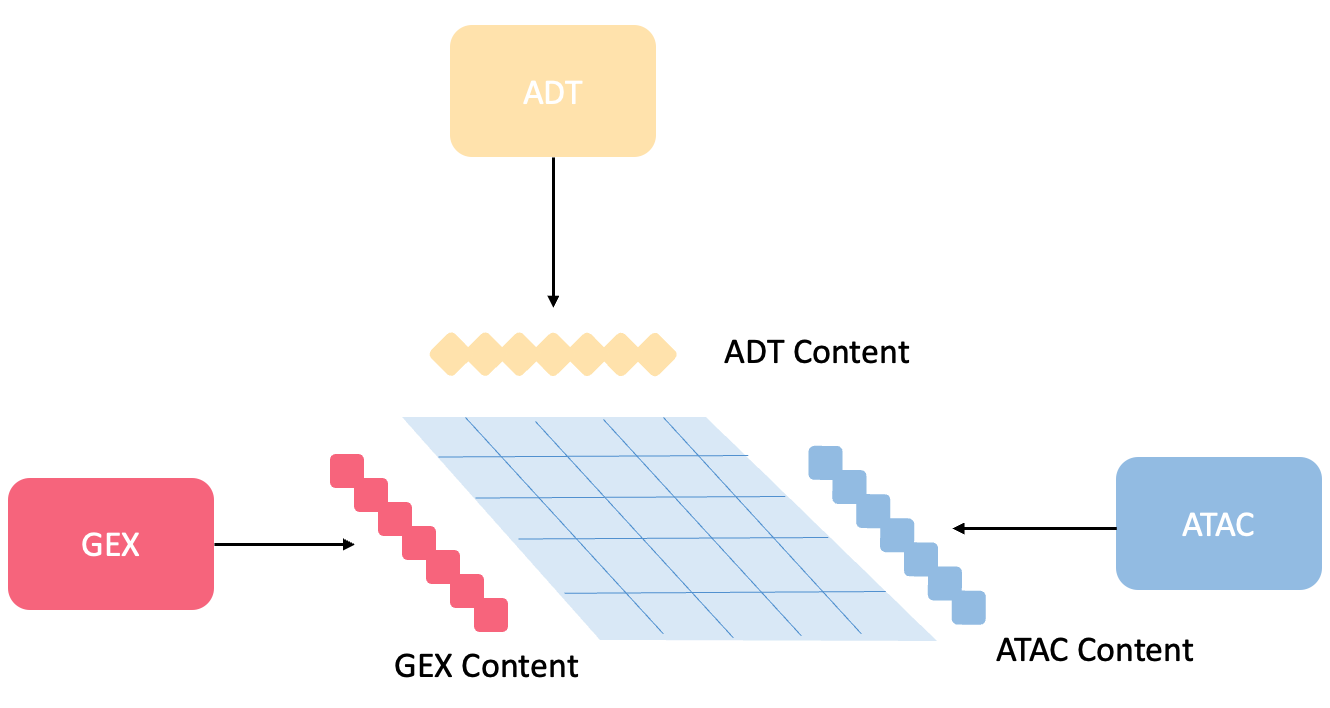
Because this research is still in-depth development, many details are being optimized. If you are interested in my research or have better ideas, we can discuss together. Please feel free to contact me at [email protected].